Feb., 2021
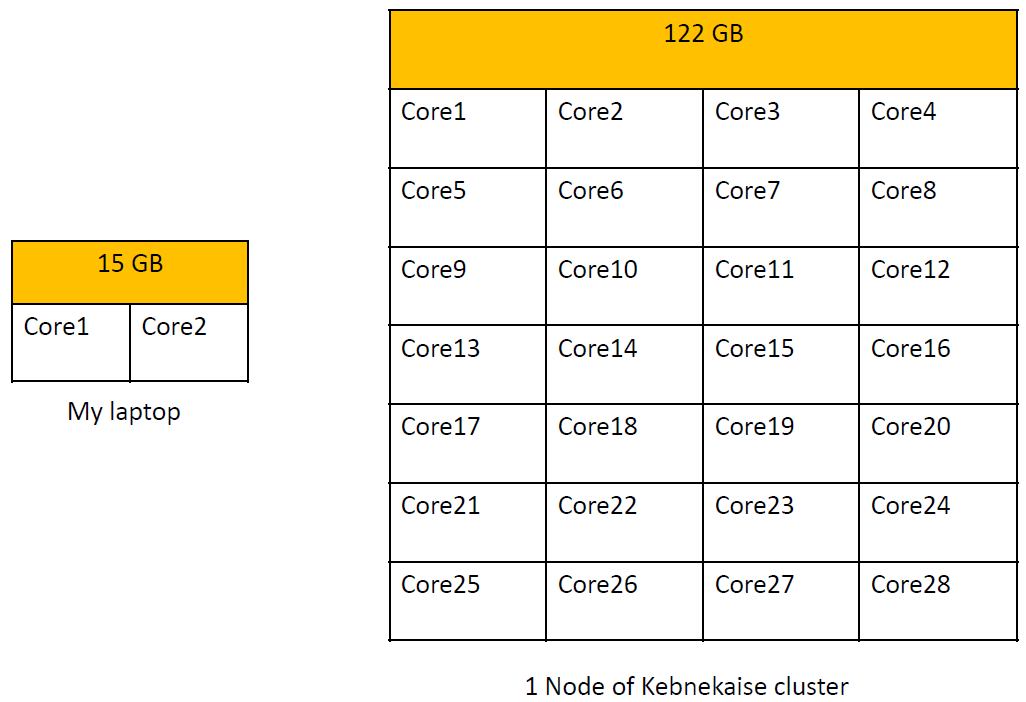
Desktop PC vs. HPC architectures

Types of programs
Programs could be of different types:
Compute bound if they use mainly CPU power (more cores can help)
Memory bound if the bottlenecks are allocating memory, copying/duplicating objects (large memory nodes on Kebnekaise)
Parallelization levels
Implicit parallelism is included in some packages, one only needs to assign the number of workers (threads)
Explicit parallelism requires the intervention of the user to write the proper parallelization instruction (Rmpi for instance)
Data dependency
This loop can be easily parallelized:
for(i in 1:100){
b[i] = 4
a[i] = 2*b[i] + 1
}
but this one cannot because it has dependency on the values of the b vector
for(i in 1:100){
b[i] = 4
a[i] = 2*b[i-1] + 1
}
Using R in HPC
There are several versions of R installed on Kebnekaise
ml spider R
# Versions:
# R/3.3.1
# R/3.4.4-X11-20180131
# R/3.5.1-Python-2.7.15
# R/3.5.1
# R/3.6.0
# R/3.6.2
# R/4.0.0
ml spider R/3.6.0 #search for the modules needed by R
# You will need to load all module(s) on any one of the lines below before the "R/3.6.0"
# GCC/8.2.0-2.31.1 OpenMPI/3.1.3
Using R in HPC
R --help #Usage: R [options] [< infile] [> outfile] # or: R CMD command [arguments] #Start R, a system for statistical computation and graphics, with the #specified options, or invoke an R tool via the 'R CMD' interface. #Options: # -h, --help Print short help message and exit # --version Print version info and exit # --encoding=ENC Specify encoding to be used for stdin # --encoding ENC # RHOME Print path to R home directory and exit # --save Do save workspace at the end of the session # --no-save Don't save it # --no-environ Don't read the site and user environment files
Adding your own packages in R
SLURM workload manager
Running serial jobs
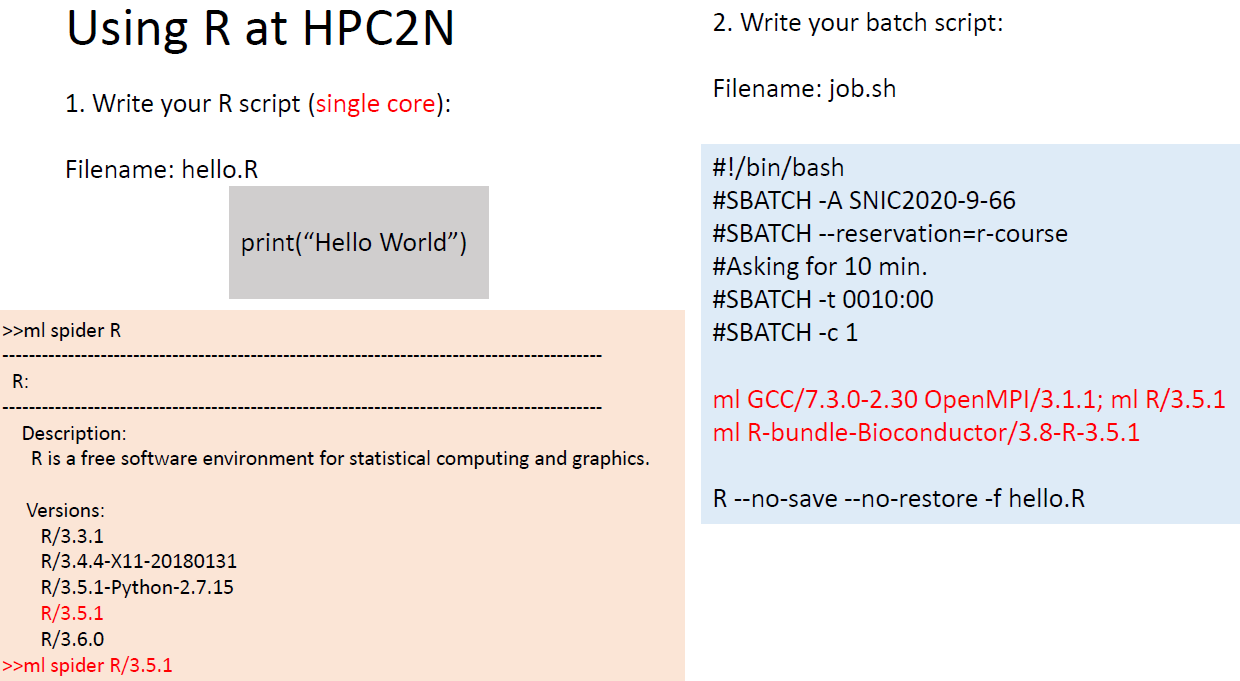
Running your script
- Transfer your files to Kebnekaise
- Submit your job with: sbatch job.sh
- In case sbatch complains about the DOS format use the command:
dos2unix job.sh
before submitting your job.
- More information: https://www.hpc2n.umu.se/resources/software/r
Running several independent jobs
One can use job arrays option in SLURM to run independent instances of a program:
#!/bin/bash #SBATCH -A Project_ID #Asking for 10 min. #SBATCH -t 00:12:00 #SBATCH --array=1-28 ##Writing the output and error files #SBATCH --output=Array_test.%A_%a.out #SBATCH --error=Array_test.%A_%a.error ml GCC/8.2.0-2.31.1 OpenMPI/3.1.3 ml R/3.6.0 R --no-save --no-restore -f script.R
Running R in parallel mode

Monitoring your jobs
squeue -a -u username list your jobs on the queue
projinfo displays the project’s usage
Parallel packages
Some packages like BLAS/LAPACK have an implicit parallelization layer that can be activated by setting a number of threads.
On Kebnekaise the OpenBLAS libraries are available and can use implicit parallelism:
sessionInfo() R version 3.6.0 (2019-04-26) Platform: x86_64-pc-linux-gnu (64-bit) Running under: Ubuntu 16.04.6 LTS Matrix products: default BLAS/LAPACK: /cvmfs/.../8.2.0-2.31.1/OpenBLAS/0.3.5/lib/libopenblas_haswellp-r0.3.5.so
Parallel packages
The number of threads can be controlled with the RhpcBLASctl package and setting the number of threads:
library(RhpcBLASctl)
n <- 5000; nsim <- 3 #matrix size nxn; nr. independent simulations
set.seed(123); summa <- 0; x <- 0; blas_set_num_threads(1) #set the number of threads
for (i in 1:nsim) {
m <- matrix(rnorm(n^2), n); a <- crossprod(m) #random matrix and symmetrize it
timing <- system.time({
x <- eigen(a, symmetric=TRUE, only.values=TRUE)$values[1] #compute eigenvalues
})[3] ; summa <- summa + timing
} ; times <- summa/nsim
cat(c("Computation of eig. random matrix 5000x5000 (sec): ", times, "\n"))
Parallel packages
Other packages (doParallel, parallel, doMC, doMPI, doFuture) use a common set of instructions to use parallel capabilities as follows:
library("package-name")
cl <- makeCluster(NumberofCores)
register_cluster(cl)
... #code to be run in parallel mode
stopCluster(cl)
Parallel packages: examples
the foreach package is used for executing loops:
library(foreach)
r <- foreach(icount(trials), .combine=cbind) %do% {
ind <- sample(100,100, replace=TRUE)
result1 <- glm(x[ind,2]~x[ind,1], family=binomial(logit))
coefficients(result1)
}
Parallel packages: examples
doParallel is a backend parallel package for executing code in parallel mode:
library(doParallel) #using "doParallel" package cl <- makeCluster(2) registerDoParallel(cl) getDoParWorkers() #this line tells the nr. of workers
## [1] 2
getDoParName() #this line tell the type of cluster
## [1] "doParallelSNOW"
stopCluster(cl)
Parallel packages: examples
doParallel can be used to execute foreach loops in parallel:
library(doParallel) #using "doParallel" package
cl <- makeCluster(2)
registerDoParallel(cl)
r <- foreach(icount(trials), .combine=cbind) %dopar% {
ind <- sample(100,100, replace=TRUE)
result1 <- glm(x[ind,2]~x[ind,1], family=binomial(logit))
coefficients(result1)
}
stopCluster(cl)
Parallel packages: examples
Send slices of data to the workers with the parallel package
library(parallel) #using "parallel" package
detectCores()
P <- detectCores(logical = FALSE) #only physical cores
myfunc <- function(id) { #function to sum by rows
arguments <- mydata[id, ]
arguments$one + arguments$two + arguments$three }
cl <- makeCluster(P) #distribute the work across cores
clusterExport(cl, "mydata")
res <- clusterApply(cl, 1:N, fun = myfunc)
stopCluster(cl)
Parallel packages: examples
The doParallel package is used in the following example to compute the eigenvalues of matrices growing in size:
library(doParallel)
my_eigen <- function(x) {
n <- x*800
m <- matrix(runif(n^2),n,n)
m[lower.tri(m)] = t(m)[lower.tri(m)]
d <- diag(eigen(m)$values)
}
cl <- makeCluster(4)
registerDoParallel(cl)
system.time( res1 <- foreach(n = 1:6) %dopar% my_eigen(n) )[3]
stopCluster(cl)
#Elapsed
#211.25
Parallel packages: examples
We can also use the future package in R which runs on several OS and supports asynchronous calculations:
library(future)
plan(multisession, gc = TRUE, workers = 4)
plan(multisession, workers = 4)
par_future <- function(x) {
#creating futures
ft <- lapply( x, function(x) future(my_eigen(x)) )
#get futures
get_ft <- lapply(ft, value)
}
x <- 1:6
system.time( res2 <- par_future(x) )[3]
#Elapsed
#210.17
Random Numbers in parallel simulations
The following simulations res1 and res2 do not give reproducible results:
library(doParallel) cl <- makeCluster(2) registerDoParallel(cl) set.seed(1) res1 <- foreach(n = rep(2, 3), .combine=rbind) %dopar% rnorm(n) set.seed(1) res2 <- foreach(n = rep(2, 3), .combine=rbind) %dopar% rnorm(n) stopCluster(cl) identical(res1,res2)
## [1] FALSE
Random Numbers in parallel simulations
For reproducible parallel simulation a RNG package such as doRNG is recommended:
library(doRNG) cl <- makeCluster(2) registerDoParallel(cl) registerDoRNG(1) res3 <- foreach(n = rep(2, 3), .combine=rbind) %dopar% rnorm(n) set.seed(1) res4 <- foreach(n = rep(2, 3), .combine=rbind) %dopar% rnorm(n) stopCluster(cl) identical(res3,res4)
## [1] TRUE
Profiling Memory: gc (Parallel)
Memory profiling is crucial upon using parallel packages. Suppose we have a data frame mydata which will be processed with the clusterApply function
gcinfo(TRUE) #activate gc N <- 5000000 mydata <- data.frame(one=1.0*seq(N),two=2.0*seq(N),three = 3.0*seq(N)) #... #Garbage collection 23 = 14+2+7 (level 0) ... #43.5 Mbytes of cons cells used (66%) #130.4 Mbytes of vectors used (65%) gc() # used (Mb) gc trigger (Mb) max used (Mb) #Ncells 572516 30.6 1233268 65.9 1233268 65.9 #Vcells 16492769 125.9 26338917 201.0 19085502 145.7
Profiling Memory: gc (Parallel)
Then, we use a function to partition the data frame by cores
library(parallel) #using parallel package
detectCores()
P <- detectCores(logical = FALSE) #only physical cores
myfunc <- function(id) { #function to sum by rows
arguments <- mydata[id, ]
arguments$one + arguments$two + arguments$three
}
Profiling Memory: gc (Parallel)
cl <- makeCluster(P) #distribute the work across cores clusterExport(cl, "mydata") res <- clusterApply(cl, 1:N, fun = myfunc) stopCluster(cl) #... #Garbage collection 1196 = 1128+50+18 (level 0) ... #312.5 Mbytes of cons cells used (60%) #206.5 Mbytes of vectors used (59%) gc() # used (Mb) gc trigger (Mb) max used (Mb) #Ncells 5850436 312.5 9776540 522.2 9776540 522.2 #Vcells 27062930 206.5 45804848 349.5 42982557 328.0
the time to execute myfunc in parallel mode increases drastically.
Good practices
- Use the login nodes for lightweight tasks
- Profile your code
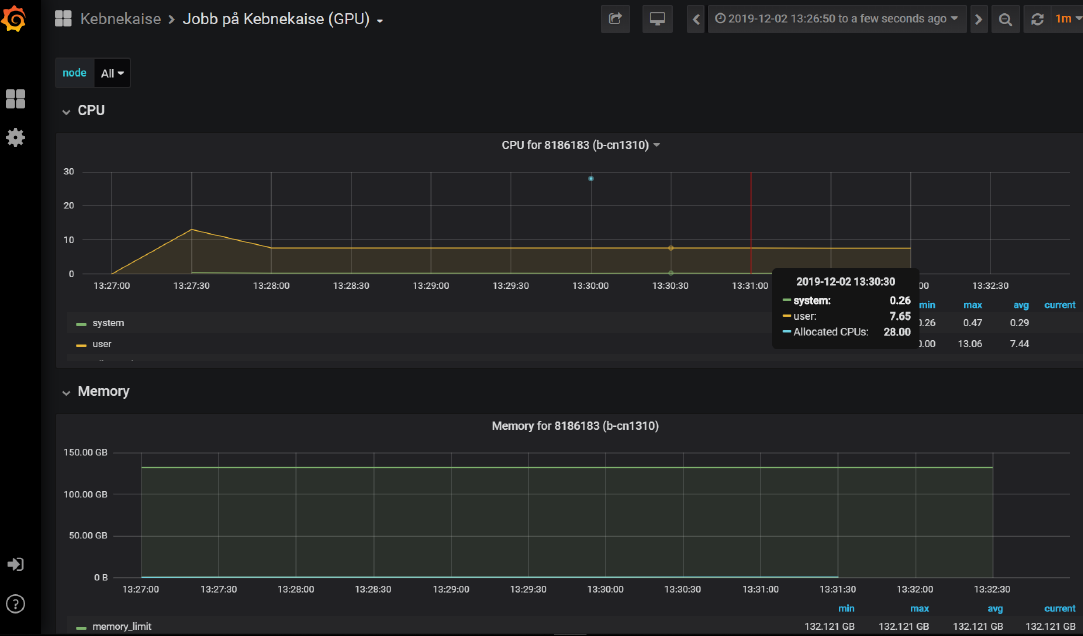
- Monitoring your job on the fly:
If you run your script on multiple cores, you can monitor the CPU and memory usage in real time, use the following command on the terminal:
job-usage “job_ID”
Then copy and paste the URL on your local web browser.
Good practices
Good practices
- If you have any issue when using R (or any software) report the case by creating a support ticket at support@hpc2n.umu.se (HPC2N users).
- It would help if you could provide a folder with the smallest example that shows the reported issue
Summary
Login nodes and Rstudio should be used for lightweight tasks for other tasks use the SLURM batch system sbatch script
Compute bound or memory bound programs
Some packages for instance the linear algebra ones include already implicit parallelism
It is a good practice to get a profiling analysis (time vs. nr. cores) to request the optimal nr. of cores in your batch script
Monitor the behavior of your batch job with job-usage tool
References
- https://swcarpentry.github.io/r-novice-inflammation/
- https://www.tutorialspoint.com/r/index.htm
- R High Performance Programming. Aloysius, Lim; William, Tjhi. Packt Publishing, 2015.
- http://adv-r.had.co.nz/memory.html
- https://blogs.oracle.com/r/managing-memory-limits-and-configuring-exadata-for-embedded-r-execution
- https://rawgit.com/PPgp/useR2017public/master/tutorial.html
- https://cran.r-project.org/web/packages/future/vignettes/future-1-overview.html